
Getting started
Klaus K. Holst and Benedikt Sommer
Source:vignettes/gettingstarted.Rmd
gettingstarted.RmdIntroduction
The carts package is a clinical trial simulation tool
for power and sample size estimation. A modular, building-block design
allows the simulation of many common clinical trial designs with only a
few lines of code. In addition, the package provides a range of
estimators for continuous, count and survival endpoints. This vignette
introduces the basic functionalities of the carts package
and aims to familarize the reader with the package interface. The
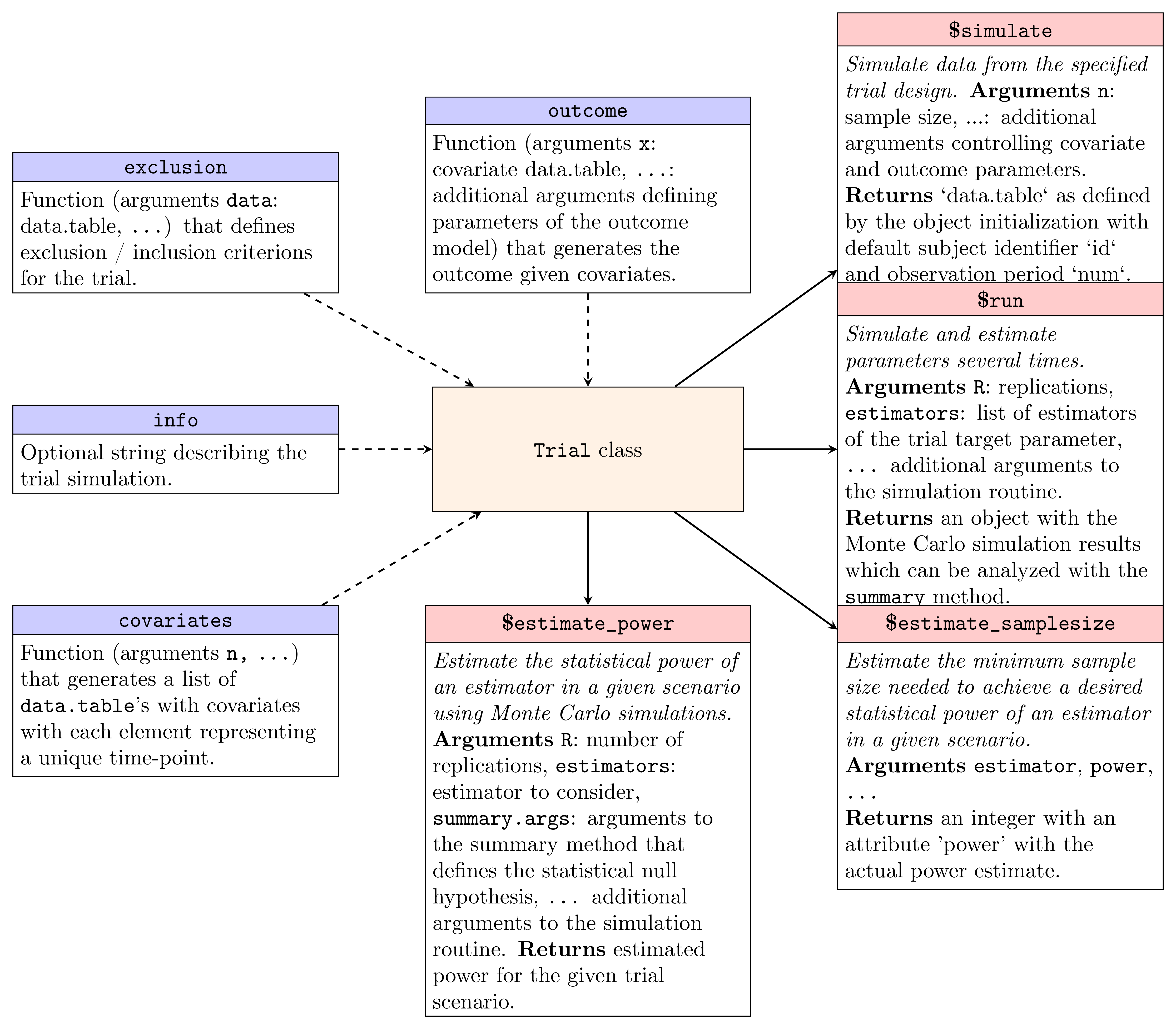
centerpiece of the interface is the R6
reference class Trial, with the constructor and main
methods visualized in Figure 1. To this end, we will consider a
two-armed parallel trial design to introduce the Trial
class and demonstrate how power and sample size estimation are carried
out conveniently.
Preliminaries
Setting up parallel computations
The carts package is basing its power and sample size
estimation on Monte Carlo integration techniques. To deal with the
computationally intensive procedures, the package uses extensive
parallel computations based on the future package. To
enable this, all that needs to be done is to define the desired parallel
backend, i.e.
This enables parallelization on all available cores on the current machine. It is equally straight-forward to set up a parallel batch computations on a cluster (see for example future.batchtools).
Progress bars and logging information
As some calculations can be time-consuming we recommend enabling progress-bars, which can be achieved with the following code.
progressr::handlers(global = TRUE)
progressr::handlers(progressr::handler_progress(times = 100))We refer to the progressr vignette for information on how to customize the progress bar appeareance.
Most output in the carts package is handled by the logger logging utility. We
refer to the following article
for information on how to alter the logging level and how to change the
destination of the log records, which can be useful when running the
carts package as batch jobs on a cluster.
Two-armed parallel design
We will consider a two-armed parallel design where the outcome of interest is with treatment variable which is independent of the baseline covariate .
Trial model
To simulate this trial we first define the random number generators for treatment indicator and baseline covariate
covariate <- function(n, p.a = 0.5) {
return(
data.frame(
a = rbinom(n, 1, p.a),
w = rnorm(n)
)
)
}which in the context of carts is a covariate model. The
general rule for simulating trial designs with time-invariant outcomes,
i.e. outcomes that are only a function of information that is available
at baseline/randomization, is that the covariate model is a function
with first argument n (sample size) that returns covariate
data in tabulated form, either as a data.frame or
data.table. Additional parameters of the covariate model
can be added after the sample size argument n, being here
the treatment probability p.a.
The trial model for this design is completed with the help of
outcome_continuous. This built-in function generates
continuous outcomes from tabulated baseline covariates, i.e. the output
of the covariate model. The default arguments of built-in functions can
be updated conveniently with setargs to the desired
parameters of the trial. Simulating the trial
requires to update the
default arguments of outcome_continuous as
outcome <- setargs(
outcome_continuous,
mean = ~ 1 + a + w,
par = c(10, -0.5, -1.2), # coef order as defined by the above formula
sd = 1.5 ** 0.5
)Finally, we instantiate a model object from the R6 Trial
class
trial <- Trial$new(
covariates = covariate,
outcome = outcome,
exclusion = identity, # no exclusion criteria (default)
info = "Two-armed parallel design" # optional info
)where, by default, no exclusion model is used.
Trial simulation
Simulating data from the defined model object is done via the simulate method
dd <- trial$simulate(n = 1e4)The method is a wrapper calling carts:::trial_simulate
which combines sampling data from the covariate and outcome model, and
the application of the exclusion model in an iterative process. First
n subjects are sampled from the covariate model. The
sampled tabulated data is then converted into a data.table
and passed on to the outcome model. The sampled covariate and outcome
data are then passed together as a single data.table to the
exclusion model. Thus, the exclusion model allows for removing subjects
based on both the covariate and outcome data. If subjects are removed,
the remaining subjects are stored and the next iteration begins by
sampling from the covariate model. The process repeats until a total of
n subjects have been sampled.
Without additional arguments, the method uses the assigned default arguments of the covariate and outcome model to generate the sample.
table(dd$a)
##
## 0 1
## 5001 4999Inspecting the simulated data
head(dd, 2)
## id a w num y
## <num> <int> <num> <num> <num>
## 1: 1 1 0.07122244 0 8.165036
## 2: 2 1 0.97029003 0 7.411990reveals that the package adds subject (id) and period
(num) identifiers to the output of the method. This is the
default behaviour that provides the functionality to follow subjects
across multiple periods in within-subject trial designs.
Updating model parameters
Additional arguments are accepted by the simulate method that allow for simulating from the same trial object while varying model parameters. This circumvents the instantiation of a new model object and provides an elegant way to carry out parameter sensitivity analyses or interactively use a trial model
trial$simulate(n = 1e4, p.a = 0.9)$a |> table()
##
## 0 1
## 1017 8983In fact, most public methods of the Trial class allow
passing arguments to the underlying covariate, outcome and exclusion
models via the optional argument of the Trial method.
Safeguarding parameter clashes
The simulate method passes all defined model parameters internally to
the covariate, outcome and exclusion models. For the given example this
means that each of the model component function calls receive
p.a = 0.9 as an optional argument. This enables updating
shared parameters in all model components with a single value
trial0 <- Trial$new(
covariates = covariate,
outcome = function(data, p.a = 0.1) rbinom(nrow(data), 1, p.a)
)
trial0$simulate(1e4)[, list(y = mean(y), a = mean(a))]
## y a
## <num> <num>
## 1: 0.1009 0.4876
trial0$simulate(1e4, p.a = 1)[, list(y = mean(y), a = mean(a))]
## y a
## <num> <num>
## 1: 1 1but also opens a pathway to accidentally overwriting default
parameters of built-in functions. A simple solution for preventing this
is achieved by using functional wrappers. This is shown here for the
covariate model, where the default value of p.a cannot be
changed dynamically anymore.
Updating default parameters of a trial model
The Trial class also allows to set and update default
arguments of covariate, outcome and exclusion models. Another way to
utilise built-in functions is therefore to initialize a trial object
as
trial <- Trial$new(
covariates = covariate,
outcome = outcome_continuous
)and then set default values for the outcome model with
trial$args_model(
mean = ~ 1 + a + w,
par = c(10, -0.5, -1.2)
)Using the args_model method updates the
model.args attribute of the trial object, which are then
used internally as the default option to simulate the trial data.
trial$simulate(1e4)[, list(y = mean(y)), by = "a"]
## a y
## <int> <num>
## 1: 0 10.037275
## 2: 1 9.515117However, the parameters in trial$model.args are
overruled when parameters with matching names are passed to the simulate
method.
Estimators
The carts package provides a range of estimators for
various trial designs. All built-in estimators are function generators
that return an estimator, which is a function of data (first input
argument). To be compatible with the package design, each estimator must
return a named vector that includes the estimate (Estimate) and standard
errors (“Std.Err”). The returned vector may include additional elements
as they not interfere with any internal procedures. A crude (marginal)
estimator can be defined by using est_glm, which is a
function generator
estimator <- est_glm(
response = "y", # default value
treatment = "a" # default value
)
data <- trial$simulate(n = 300)
estimator(data)
## Estimate Std.Err 2.5% 97.5% P-value
## a -0.5394 0.1918 -0.9153 -0.1635 0.004914Estimators can be assigned to a trial object either during its
initialization via the estimators argument of
Trial$new or after initialization
trial$estimators(marginal = est_glm()) # using def. args. of est_glmand all assigned estimators can be retrieved by calling the estimator method
names(trial$estimators())
## [1] "marginal"or printing the trial object
print(trial)
## ── Trial Object ──
##
## Model arguments:
## • mean: Class 'formula' language ~1 + a + w
## • par: num [1:3] 10 -0.5 -1.2
##
## Summary arguments:
## • level: num 0.05
## • null: num 0
## • ni.margin: NULL
## • alternative: chr "!="
## • reject.function: NULL
## • true.value: NULL
## • nominal.coverage: num 0.9
##
## Covariates:
## function (n, p.a = 0.5, ...)
##
## Outcome:
## function (data, mean = NULL, par = NULL, sd = 1, het = 0, outcome.name = "y", remove = c("id", "num"), family = gaussian(), ...)
##
## Exclusion:
## function (x, ...)
##
## Estimators:
## 1. marginal
## ────────────────────Power estimation
Carrying out power estimations is straightforward once estimators are assigned to the trial model.
trial$estimate_power(
n = 300, # sample size
R = 500 # number of Monte Carlo replicates
)
## [1] 0.778Hypothesis testing
The method uses carts:::summary.runtrials internally to
implement the hypothesis test for a given trial design. By default, a
two-sided test is carried out for the null hypothesis of no treatment
effect with 0.05 signifcance level. The test can be changed via the
summary.args argument
trial$estimate_power(n = 300, R = 500,
summary.args = list(alternative = ">") # one-sided test should
# have no power as a < 0
)
## [1] 0Updating model parameters
Arguments to the covariate, outcome and exclusion models can be
passed via the optional arguments of the method. The behavior is
identical to the one of Trial$simulate. That is, passed
arguments overwrite their default values in
Trial$model.args.
trial$estimate_power(n = 300, R = 500, p.a = 0.1)
## [1] 0.376One should note that the current interface does not pass on these optional arguments to estimators. As a consequence, all estimators must be initialized with values assigned to their optional arguments.
Multiple estimators
The default settings of carts is to estimate the power
of all estimators that are defined in Trial$estimators.
trial$estimators(adjusted = est_glm(covariates = "w"))
trial$estimate_power(n = 300, R = 500)
## marginal adjusted
## 0.792 0.992Sample-size estimations
Last, we estimate the sample size required to attain 90% power for the specified trial model and estimator
# trial$estimate_samplesize(
# power = 0.9, # default
# estimator = trial$estimators("adjusted")
# )The method optimizes the sample size by internally calling the
Trial$estimate_power method. Therefore, the behavior for
updating model parameters and changing the default settings for the
hypothesis test are identical. That is, model parameters are passed as
optional arguments or set via the args_model method, and
the summary.args argument is used to change the hypothesis
test.